The coralligenous environment is a typical Mediterranean marine habitat and hosts the highest concentration of marine biodiversity in the Med. Paradoxically, unlike Posidonia meadows, it is poorly studied. Like the famous tropical coral reefs, it is a complex biogenic habitat, based on encrusting red algae (numerous metazoan phyla also participate to bio-construction). This project focuses on understanding its functioning and resilience capacity, and on establishing biodiversity and local and ecological connectivity patterns, on providing information for rationalizing the design of Marine Protected Area networks and on monitoring methods.
The project relies on interdisciplinarity, combining population genetics, community ecology, microbial ecology and physical oceanography. Its tests hypotheses related to the niche and neutral biodiversity theories that include both inter- and intra-specific levels of biodiversity. The societal interest relies on the setup of innovative, cost-effective and accurate methods for biodiversity characterization and monitoring, based on genetic tools rigorously inter-calibrated with traditional taxonomy and photo-quadrate approaches. This requests four tasks, partly using the same data, but different analysis methods:
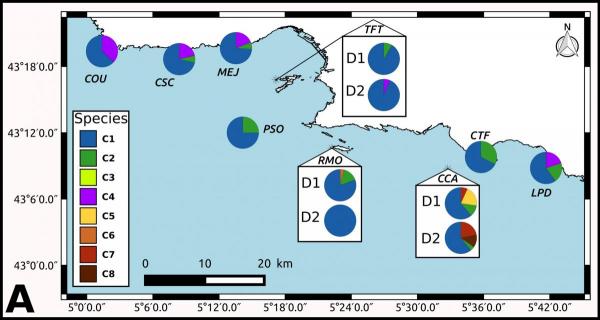
1- Establishing the taxonomic composition of the engineer corallinale algae.
2- Establishing the species composition for numerous small quadrates of coralligenous using meta-barcoding in different ecological profiles.
3- Population structure and phylogeography of two selected taxa: a red alga Lithophyllum spp. and a bryozoan Myriapora truncata,
4- Comparing intra-specific diversity among species (determine site-effects vs species-effects); species assemblages ; intra- and inter-specific diversity (including microbial); how these coralligenous data support niche vs neutral biodiversity theories;
5 - Proposing new GES indicators (good environmental status) for the coralligenous and biodiversity management rules.